# install.packages("tidyverse")
library(tidyverse)Introduction to tidyverse
The R Tidyverse is a collection of packages for data handling, analysis and visualization. If you want to use the tidyverse, you have to install the additional packages first with the install.packages() function. Once installed, you then have to tell R to make the tidyverse functions available in your current R session with library() You only have to install a package once, but loading it has to be done every time you start a new R session. It is recommened to either not include the install.packages() in your script or just comment it out like below.
As you see in the output, library(tidyverse) actually loads nine different packages. It will also give you a warning about conflicting functions. Do not worry for now, we will get to that in time.
Why tidyverse?
- consistent syntax and workflows
- makes code more readable
- pipe operator
%>%/|>can chain functions together - tidy data approach
- rows are observations
- columns are variables / features
data.frames with dplyr
- provides functions for
data.framemanipulation - can complement or replace base R functions
Of course, you can also load single packages from the tidyverse with the library() function.
library(dplyr)
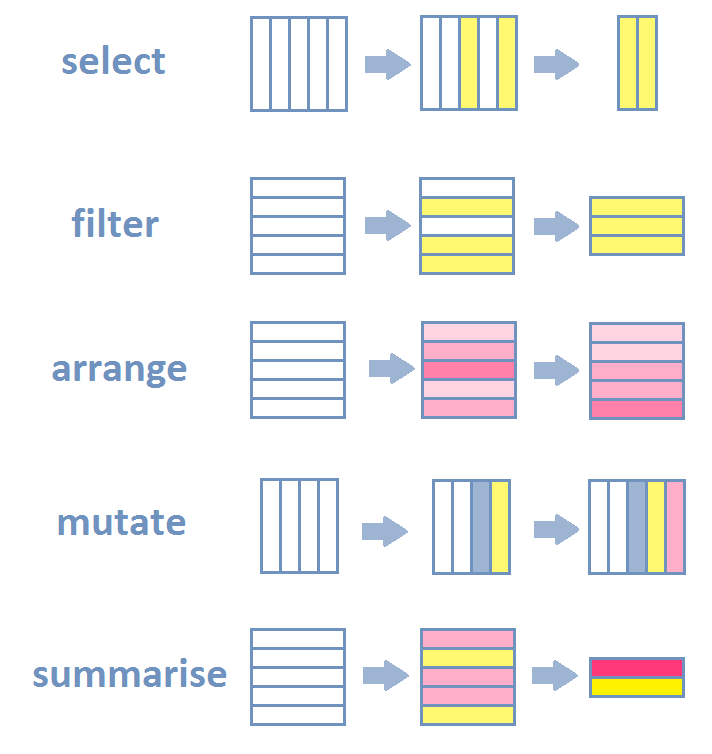
trees = read.csv("data/muenster_trees.csv")slice - a slice of data - i.e. the specified rows
trees = slice(trees, seq(8))select - selects columns
select(trees, species) species
1 Tilia
2 Tilia
3 Carpinus
4 Carpinus
5 Carpinus
6 Tilia
7 Carpinus
8 Platanusfilter - filters rows based on logical operators
filter(trees, species == "Tilia") species district X Y
1 Tilia Uppenberg 404685.2 5759126
2 Tilia Uppenberg 404698.5 5759130
3 Tilia Uppenberg 404685.6 5759520mutate - mutates the data.frame by adding columns
mutate(trees, city = "Muenster") species district X Y city
1 Tilia Uppenberg 404685.2 5759126 Muenster
2 Tilia Uppenberg 404698.5 5759130 Muenster
3 Carpinus Uppenberg 404693.0 5759279 Muenster
4 Carpinus Uppenberg 404692.9 5759289 Muenster
5 Carpinus Uppenberg 404692.1 5759319 Muenster
6 Tilia Uppenberg 404685.6 5759520 Muenster
7 Carpinus Uppenberg 404662.3 5760350 Muenster
8 Platanus Neutor 404352.7 5758912 Muenstersummarise - summarises data
summarise(trees, amount = n_distinct(species)) amount
1 3pull - pulls the values out of a column
pull(trees, species)[1] "Tilia" "Tilia" "Carpinus" "Carpinus" "Carpinus" "Tilia" "Carpinus"
[8] "Platanus"Note that the functions above could all be realized with base R. Think about the tidyverse as a different dialect to data analysis with R. Later on, it will be up to you which style you like more and feels more natural to your thought process. However, for understanding code you randomly find on the internet or if you work with other people that prefer different dialects than yourself, you should be able to read and write the basics of each style regardless.
# the same in base R
# select
trees$species
# filter
trees[,trees$species == "Tilia"]
# mutate
trees$city = "Muenster"
# summarise
length(unique(trees$species))The pipe operator
The strength of dplyr is the possibility to chain functions with %>% or |>.
trees|>
filter(species == "Tilia" | species == "Platanus") |>
pull(district) |>
unique()[1] "Uppenberg" "Neutor" With base R functions this looks messy, because we have to use functions inside functions.
unique(trees$district[trees$species == "Tilia" | trees$species == "Platanus"])[1] "Uppenberg" "Neutor"